ggplot2 Tutorial: Components, Layers, and Examples
Nikhil Gopal, Graham Kim, Uba Backonja
January 25th, 2016
Goal of this presentation
- Learn to deconstruct an image
- Learn to understand how to read ggplot code
- Understand how ggplot constructs your code into a figure
What you need to follow along
- R (DSL): brew install homebrew/science/r
- RStudio (IDE): brew install Caskroom/cask/rstudio
- Assumes you have homebrew installed (if you don't, you should!)
- ggplot2 (library) and HSAUR3 (library)
install.packages("ggplot2")
install.packages("HSAUR3")
The BP dataset
A data frame with 53 observations on the following 3 variables.
- logdose = the logarithm (base 10) of the dose of drug in milligrams
- bloodp = average systolic blood pressure achieved while the drug was being administered
- recovtime = time in minutes before the patient's systolic blood pressure returned to 100mm of mercury
A peek into the data frame
require(HSAUR3)
head(bp)
logdose bloodp recovtime
1 2.26 66 7
2 1.81 52 10
3 1.78 72 18
4 1.54 67 NA
5 2.06 69 10
6 1.74 71 13
Let's Summarize the BP dataset
When your data is in the right data structure, summarizing your data is cake!:
summary(bp)
logdose bloodp recovtime
Min. :1.180 Min. :51.00 Min. : 4.0
1st Qu.:1.740 1st Qu.:60.00 1st Qu.:10.5
Median :1.900 Median :67.00 Median :21.0
Mean :1.992 Mean :66.34 Mean :22.4
3rd Qu.:2.260 3rd Qu.:69.00 3rd Qu.:26.5
Max. :2.780 Max. :88.00 Max. :72.0
NA's :10
Visualizing data in R using defaults
In R, graphing data is easy!
require(HSAUR3)
plot(bp)
Visualizing data in R using defaults
plot(bp)
But then why ggplot2?
- Default graphics in R is easy, but less customizable
- Default graphics in R is easy, but figures can be made more domain specific
- Default graphics in R is easy, but arguably less aesthetic
What is ggplot2?
ggplot2 is a visualization library based on the Grammar of Graphics (by Leland Wilkinson, who is now at Tableau).
In short, it is the hypothesis that all graphics can be composed from the same material parts.
What components underlie all graphics?
- Geoms (used to represent data points)
- Aesthetics (used to customize the “look” of data points)
- Stats (used to transform data)
- Scales (controls how data values relate to visual values)
- Coordinate System (coordinate plane)
- Position Adjustments (arrangement of geoms)
- Labels
- Facets (dividing a plot into subplots)
- Legends
- Zooming
- Themes (color schemes, University of Washington colors, etc.)
What we will cover today
- Geoms (used to represent data points)
- Aesthetics (used to customize the “look” of data points)
- Stats (used to transform data)
- Scales (controls how data values relate to visual values)
- Coordinate System (coordinate plane)
- Position Adjustments (arrangement of geoms)
- Labels
- Facets (dividing a plot into subplots)
- Legends
- Zooming
- Themes (color schemes, University of Washington colors, etc.)
BP in default graphics (again)
Let's focus in on logdose vs bloodp
Let's focus in on logdose vs bloodp
- Now, let's deconstruct!
Deconstruction of a figure
- geom = circles/points
- coordinate system = cartesian
- X scale = continuous
- Y scale = continuous
Let's reconstruct using ggplot2
require(ggplot2)
ggplot(bp, aes(logdose, bloodp)) +
geom_point() +
coord_cartesian()
Let's reconstruct using ggplot2
Understanding the syntax
require(ggplot2)
ggplot(bp, aes(logdose, bloodp)) +
geom_point() +
coord_cartesian()
- “require” loads the ggplot2 library. You can also use “library”
- “bp” refers to the dataset (it must be a data frame)
- “aes” refers to aesthetics (what we attach visual encodings to)
- “+” tells ggplot2 that we are about to add a layer
- “geom_point()” is a function that adds a points layer
- “coord_cartesian()” is a function that adds a coordinate layer
- We will refer back to this “stub” code several times later in the presentation
Layers
What happens if we just run
ggplot(bp, aes(logdose, bloodp))
without any layers?:
Layers
What happens if we just run
ggplot(bp, aes(logdose, bloodp))
without any layers?:
Error: No layers in plot
Execution halted
Let's pause and reflect for a moment
- ggplot2 operates via layers, and needs at least one to display something
- layers are additive (and there are defaults)
- ggplot2 requires datasets to be data frames
- foundational idea: every figure can be composed using a combination of core components
- visual encoding assignments happen in
aes, but encoding details are provided in layers
Let's modify our plot
Can anyone describe how they might tweak the previous code to create this plot?
Let's modify our plot
Previous code:
ggplot(bp, aes(logdose, bloodp)) +
geom_point() +
coord_cartesian()
Let's modify our plot
If you deconstruct the plot, what is the main difference between the original plot and this one? The coordinate system!
ggplot(bp, aes(logdose, bloodp)) +
geom_point() +
coord_polar()
Let's add color!
What do you see? Can anyone describe how they might create this plot? What would you need to add to the stub code and where would you add it?
Let's add color!
logdose bloodp recovtime
1 2.26 66 7
2 1.81 52 10
3 1.78 72 18
4 1.54 67 NA
5 2.06 69 10
6 1.74 71 13
ggplot(bp,
aes(logdose, bloodp, color=recovtime)
) +
geom_point() +
scale_color_continuous(
low="#FFFFFF",
high="#990000"
)
Color choices: RColorBrewer
Let's add size!
What do you see? Can anyone describe how they might create this plot?
Let's add size!
What do you see? Can anyone describe how they might create this plot?
logdose bloodp recovtime
1 2.26 66 7
2 1.81 52 10
ggplot(bp,
aes(logdose, bloodp, color=recovtime, size=recovtime)
) +
geom_point() +
scale_color_continuous(
low="#FFFFFF",
high="#990000"
)
And shapes!
Do you remember anything tricky about shapes from previous lectures? What kind of scale is shape best used for? And what kind of scale do we have in our dataset?
And shapes!
I don't think you'd want to map shapes in this way. I've demonstrated this solely as an excuse to use the shape encoding!
ggplot(bp,
aes(logdose, bloodp, color=recovtime, shape=factor(bp$recovtime > 50), size=recovtime)
) +
geom_point() +
scale_color_continuous(
low="#FFFFFF",
high="#990000"
)
And a quick linear regression line
Can anyone describe how they might create this plot? What do you see?
And a quick linear regression line
Can anyone describe how they might create this plot? What do you see?
ggplot(bp,
aes(logdose, bloodp, color=recovtime, size=recovtime)
) +
geom_point() +
geom_smooth(method=lm) +
theme(legend.position="none") +
scale_color_continuous(
low="#FFFFFF",
high="#990000"
)
Titles and Labels
Always have titles and labels!
Titles and Labels
Can anyone describe how they might create this plot? What do you see?
- I like when figures titles describe the relationship or items that are being called to attention (not a hard and fast rule, likely more my own style)
Titles and Labels
ggplot(bp,
aes(logdose, bloodp)
) +
geom_point() +
coord_cartesian() +
ggtitle("log10-transformed hypotensive agent dosage \npositively correlated with\naverage systolic blood pressure (while drug was being administered)") +
theme(plot.title = element_text(
lineheight=.8,
face="bold")
)
Let's meditate on this for a second...
- The examples we just walked through were designed to illustrate how to call and use layers in ggplot2
- Double and triple encoding the same groups of data can facilitate improved perceptual performance. However, there are diminishing returns and figure can end up looking “busy”.
How might we make a histogram?
- How many dimensions? Which?
- Which geom?
- Which scale?
How might we make a histogram?
How might we make a histogram?
ggplot(bp,
aes(logdose)
) +
geom_histogram(
binwidth= 0.5
)
How might we add color?
- Are there any transformations or operations we need to compute?
Let's add some color!
How might we make a histogram?
ggplot(bp,
aes(logdose,
fill=cut(
bp$logdose,
breaks = c(1,1.5,2,2.5,3)
)
)
) +
geom_histogram(
binwidth= 0.5
)
Let's make one of those fancy coxcombs we see all over the Internet!
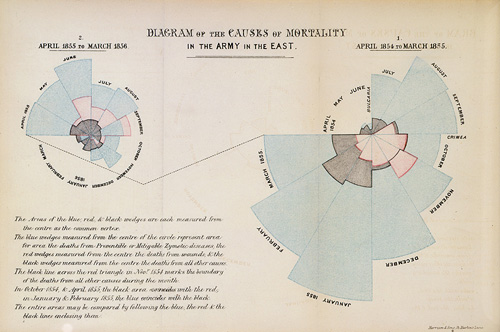
Let's make one of those fancy coxcombs we see all over the Internet!
Let's make one of those fancy coxcombs we see all over the Internet!
ggplot(bp,
aes(logdose,
fill=cut(
bp$logdose,
breaks = c(1,1.5,2,2.5,3)
)
)
) +
geom_histogram(
binwidth= 0.5
) +
coord_polar()
Which would you use, and when?
- Which figure is more perceptually accurate?
- Which figure “scales” better?
- In which situations would you use each?