How to Mine 23andMe Data: Part 3
A Note on Choice of Language:
I'm going to cheat a little bit. Taking my own advice from my post, "Bioinformatics Programming Like Experts," I've found it much simpler to answer my next few questions using R. R has a number of complicated statistical tests built-in -- performing them on data is trivial.
What I've Done: Principal Component Analysis:
I've performed principal component analysis on my family's 23andMe data. In a nutshell, principal component analysis transforms multi-dimensional data into a number of components which reflect the amount of variance in each dimension. Thus, the first principal component corresponds to the dimension which accounts for most of the variation in a dataset and the last principal component corresponds to the dimension which accounts for the least variation in a dataset. The process of obtaining these numbers is very involved.
What Are We Looking At?:
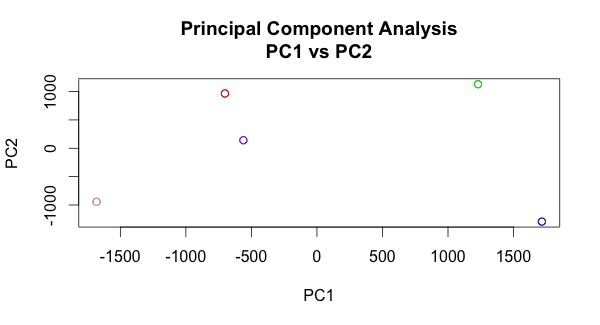
To get to the punchline and share what I've posted in simple terms, we can plot "principal component 1" values against "principal component 2" values and the similar data points will "cluster."
Since I didn't include a legend in the image above, here is who each data point corresponds to (and rough coordinates for folks who can't see color too well):
- Red = Me (-700, 1000)
- Purple = Sister (-550, 100)
- Pink = Mom (-1750, -1000)
- Green = Dad (1300, 1000)
- Blue = Grandfather (1700, -1500)